UMD vs. COVID-19
Finding a COVID-19 Drug that Works
"With COVID-19, you kind of have to do everything—the entirety of a clinical development that would take usually five, six or seven years—in two or three months…The team here was optimistic going in, knowing what we knew about the medicine and its ability to inhibit the virus, and we felt a real responsibility to try and move as quickly as we could."
Gilead Sciences' Chief Medical Officer Merdad Parsey (B.S. '85, biochemistry and microbiology) on the all-out effort to develop and test the company's COVID-19 antiviral drug remdesivir.
Learn more: go.umd.edu/covid-drug
Teaming Up to Fight COVID-19
Assistant Professor Margaret Scull and Professor Jeffrey DeStefano from UMD's Department of Cell Biology and Molecular Genetics teamed up to develop new drugs to treat COVID-19, supported by a Coronavirus Research Seed Fund Award from UMD's Division of Research. DeStefano is developing small molecules called aptamers that can bind to coronavirus surface proteins and block the virus from entering cells. Scull will screen these aptamers on a complex cell model her lab developed of the human airway—a primary target of the coronavirus—to see whether they're effective in blocking infection in airway epithelial cells.
Exploring COVID-19's Silent Spread
COVID-19's rapid worldwide spread has been fueled in part by the virus's ability to be transmitted by people who don't have any apparent symptoms. A team of researchers at Princeton University that includes Simon Levin (Ph.D. '64, mathematics) published a study that found that this stealth phase of transmission can be a successful evolutionary strategy for viruses like COVID-19. Understanding just how much asymptomatic people are contributing to the spread of the coronavirus could guide public health managers as they take steps to prevent transmission, from stay-at-home orders to social distancing.
Predicting COVID-19 via Satellite
Rita Colwell, Distinguished University Professor in the University of Maryland Institute for Advanced Computer Studies, developed a predictive model for SARS-CoV-2, the strain of coronavirus responsible for COVID-19. Colwell's team found that satellite data—including population movement, weather conditions, sea-surface heights and temperature—could help identify specific regions that may be at high risk for a future COVID-19 outbreak, making satellites a valuable predictive tool for public health.
Sharing Faces of the Pandemic
Seattle physician Jessica Lu (B.S. '14, biological sciences) teamed up with a colleague to launch an Instagram account called Frontline COVID-19. The account amplifies the voices of medical professionals on the frontlines of the fight against COVID-19 and provides a place to share their stories and pictures. For the public, it's a behind-the-scenes look at what's going on in hospitals around the world as the outbreak continues.
Creating a Popular COVID-19 Screening Tool
Adeel Malik (B.S. '15, biological sciences; B.S. '15, finance) and Bilal Naved (B.S. '15, bioengineering) created an online coronavirus symptom checker called Clearstep that is being used across the country to screen patients for COVID-19 symptoms. Users answer multiple-choice questions for symptoms of COVID-19 and 500 other health issues to determine whether they need immediate medical attention. Malik and Naved believe that as telemedicine becomes more mainstream, tools like Clearstep will be the new normal in health care.
Covering COVID-19 News
"When I wrote about the Spanish flu, I never ever thought I'd see people around the world wearing masks. I never thought society would shut down like it has and that schools would close for months."
Gina Kolata (B.S. '69, microbiology; M.A. '73, mathematics), New York Times science and medical reporter and author of "Flu: The Story of the Great Influenza Pandemic of 1918 and the Search for the Virus That Caused It," on the challenges of covering the COVID-19 pandemic.
Unraveling the Genomic Mysteries of CoVID-19
<
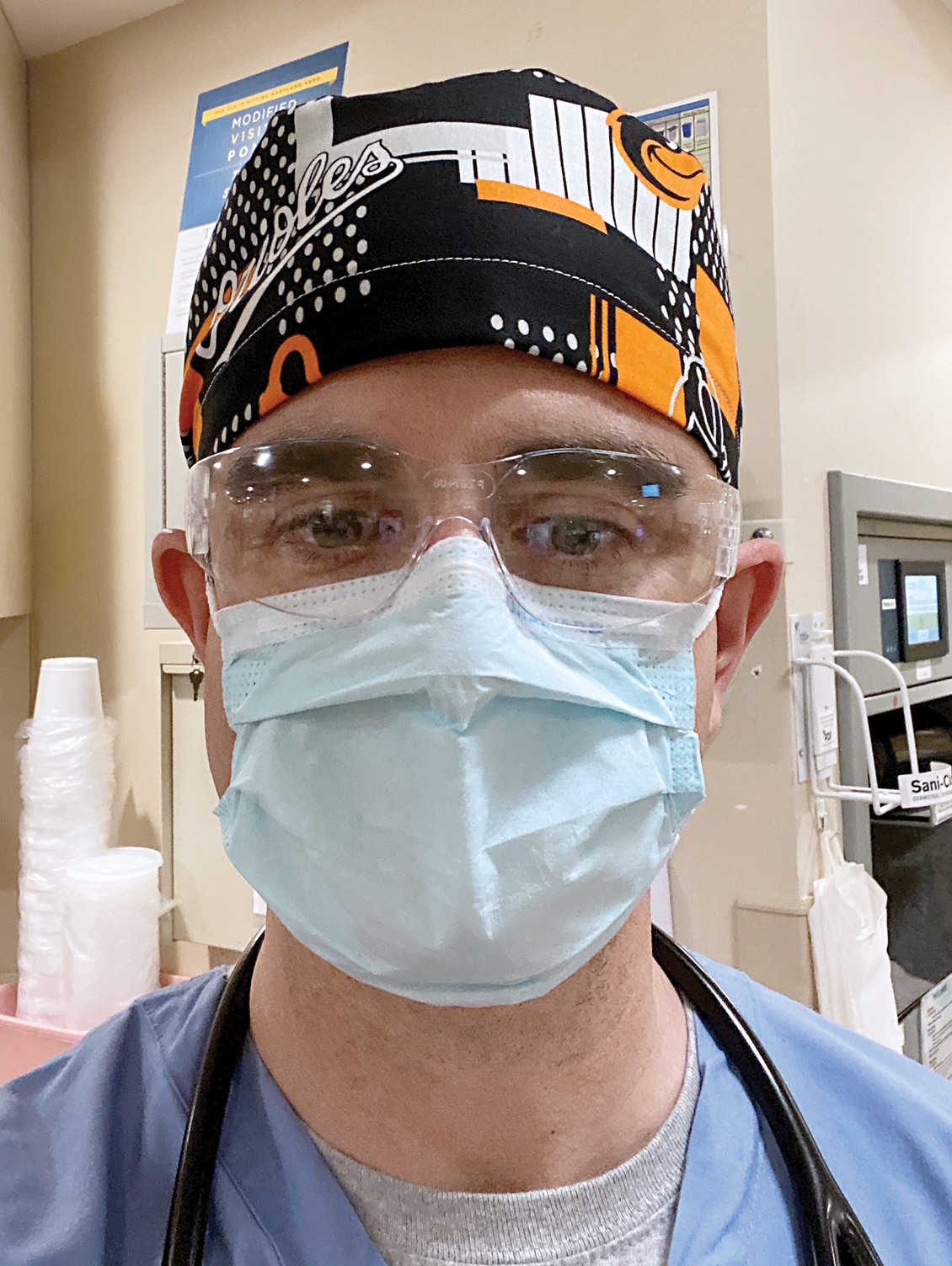
Biology Professor Michael Cummings received funding from the National Science Foundation to develop new tools and strategies for analyzing genomic data from SARS-CoV-2, the strain of coronavirus responsible for COVID-19. Understanding the genomics of COVID-19 could help scientists track its history and mutations and help public health officials predict where specific strains of the virus might pop up next.
Fighting COVID-19 in the ER
"It certainly is scary not knowing what could happen on a given day...You just take it one day at a time and try to get through. You hope you don't make any mistakes at work that would cause you to contaminate yourself."
Emergency physician Larry Edelman (B.S. '01, biochemistry) on the fears and challenges of fighting COVID-19 in the ER at Baltimore-area hospitals.
Tracking COVID-19's Impact on Air Pollution

When COVID-19 stay-at-home orders interrupted normal commutes in the D.C. metro area, a unique opportunity arose to study the impact of reduced traffic on air quality. Since March, Atmospheric and Oceanic Science Professor Russell Dickerson and his research team have been working with the National Oceanic and Atmospheric Administration, conducting aerial surveys to measure pollutants and greenhouse gases. Preliminary results show significant differences in air pollution levels before and after the lockdown.
Mapping COVID-19 in Real Time

Distinguished University Professor of Computer ScienceHanan Samet led a team in developing a novel web application that can track the progression of COVID-19 over space and time. The web application, called NewsStand CoronaViz, is a research prototype that maps COVID-19 outbreaks, based on key variables including the number of infections, active cases, recoveries and deaths that are reported daily in real time from news articles and other sources.
Battling COVID-19—And Not Just in the Lab
Jonathan Dinman, professor and chair of UMD's Department of Cell Biology and Molecular Genetics, received a Coronavirus Research Seed Fund Award from UMD's Division of Research in April to focus his research on COVID-19, specifically on a viral process called programmed-1 ribosomal frameshifting (-1 PRF), which presents a promising target for antiviral drugs. In an unexpected twist of fate, Dinman's COVID-19 research was interrupted a few weeks later when he got the coronavirus himself. "The irony of the virologist being infected with COVID-19 does not escape me, that's for sure," Dinman said.
Predicting Pandemics with the Power of Computers Computer Science
Assistant Professor Abhinav Bhatele, Computer Science Distinguished University Professor Aravind Srinivasan and Distinguished University Professor Rita Colwell—who all have appointments in the University of Maryland Institute for Advanced Computer Studies—are collaborating with scientists from across the U.S. to deploy the latest advances in artificial intelligence, machine learning, supercomputing and social science data to fight pandemics. Funded by a $10 million Expeditions grant from the National Science Foundation, the researchers are using these powerful technologies to explore factors that increase the risk of global pandemics like COVID-19, such as trends in globalization, antimicrobial resistance, urbanization and ecological pressures.
Written by Leslie Miller and Abby Robinson
This article was published in the Summer 2020 issue of Odyssey magazine. To read other stories from that issue, please visit go.umd.edu/odyssey.