UMD Researchers Study the Intricate Processes Underpinning Gene Expression
Their research reveals the dynamics of an ever-moving polymer called chromatin.
A new study led by University of Maryland physicists sheds light on the cellular processes that regulate genes. Published in the journal Science Advances, the paper explains how the dynamics of a polymer called chromatin—the structure into which DNA is packaged—regulate gene expression.
Through the use of machine learning and statistical algorithms, a research team led by Physics Professor Arpita Upadhyaya and National Institutes of Health Senior Investigator Gordon Hager discovered that chromatin can switch between a lower and higher mobility state within seconds. The team found that the extent to which chromatin moves inside cells is an overlooked but important process, with the lower mobility state being linked to gene expression.
Notably, transcription factors (TFs)—proteins that bind specific DNA sequences within the chromatin polymer and turn genes on or off—exhibit the same mobility as that of the piece of chromatin they are bound to. In their study, the researchers analyzed a group of TFs called nuclear receptors, which are targeted by drugs that treat a variety of diseases and conditions.
“The nuclear receptors in our study are important therapeutic targets for breast cancer, prostate cancer and diabetes,” explained the study’s first author, Kaustubh Wagh (Ph.D. ’23, physics). “Understanding their basic mechanism of action is essential to establish a baseline for how these proteins function.”
As a result, these findings could have broad applications in medicine.
On the move
The genetic information that children inherit from their parents is contained in DNA—the set of instructions for all possible proteins that cells can make. A DNA molecule is about 2 meters in length when stretched from end to end, and it must be compacted 100,000 times in a highly organized manner to fit inside a cell’s nucleus. To achieve this, DNA is packaged into chromatin in the nucleus of a cell, but that bundle of genetic material doesn’t stay stationary.
“We know that how the genome is organized in the nucleus of our cells has profound consequences for gene expression,” Wagh said. “However, an often-overlooked fact is that chromatin is constantly moving around inside the cell, and this mobility may have important consequences for gene regulation.”
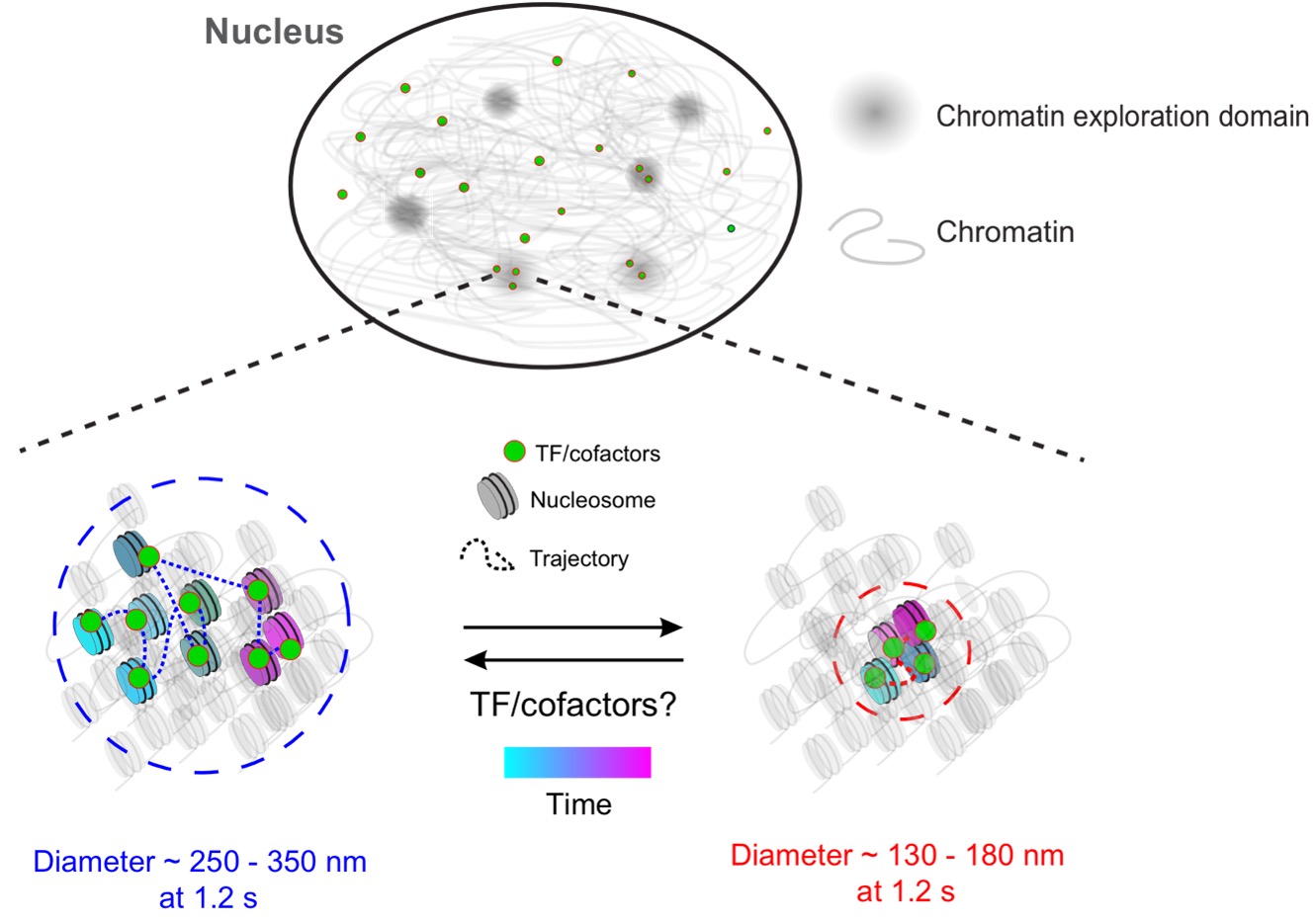
The research team—including collaborators from the National Cancer Institute, the University of Buenos Aires and the University of Southern Denmark—showed that chromatin switches between two distinct mobility states: a lower one (state 1) and a higher one (state 2). Earlier theories suggested that different parts of the nucleus had fixed chromatin mobilities, but the researchers demonstrated that chromatin is much more dynamic.
“Previous studies have proposed that different chromatin mobility states occupy distinct regions of the cell nucleus. However, these studies were performed on a sub-second timescale,” said Upadhyaya, who holds a joint appointment in the Institute for Physical Science and Technology. “We extend this model by showing that on longer timescales, the chromatin polymer can locally switch between two mobility states.”
The researchers found that transcriptionally active TFs preferred to bind to chromatin in state 1. They were also surprised to discover that TF molecules in a lower mobility state bound for longer periods of time, likely affecting gene regulation.
Finding a raft in the ocean
This study advances scientists’ understanding of chromatin dynamics and gene expression. The researchers will use their framework to study how mutations affect the function of TFs, which can offer insight into the onset of various diseases.
“We are now in a position to answer whether a particular disease phenotype occurs due to the TF binding for too much or too little time, or not binding in the right chromatin state,” Wagh said.
The team also plans to investigate how TFs achieve the challenging feat of finding their targets. TFs target a specific base pair sequence of DNA, and only by finding and binding this sequence can they recruit other proteins to activate nearby genes.
“A TF finding its target site is like finding a single raft in the middle of the ocean,” Upadhyaya said. “It’s a miracle it even happens, and we plan to figure out how.”
###
Their paper, “Dynamic switching of transcriptional regulators between two distinct low-mobility chromatin states,” was published in Science Advances on June 14, 2023.
This work was supported by the National Institutes of Health (Award No. R35 GM145313), National Cancer Institute Intramural Program, NCI-UMD Partnership for Integrative Cancer Research, Center for Cancer Research, National Science Foundation (Award Nos. NSF MCB 2132922 and NSF PHY 1915534), Vissing Foundation, William Demant Foundation, Knud Højgaard Foundation, Frimodt-Heineke Foundation, Director Ib Henriksen Foundation, Ove and Edith Buhl Olesen Memorial Foundation, Academy of Finland, Cancer Foundation Finland, Sigrid Jusélius Foundation, Villum Foundation (Award No. 73288), Independent Research Fund Denmark (Award No. 12-125524), Danish National Research Foundation (Award No. 141) to the Center for Functional Genomics and Tissue Plasticity, CONICET and the Agencia Nacional de Programación Científica y Tecnológica (Award Nos. 2019-0397 and PICT 2018-0573). This story does not necessarily reflect the views of these organizations.
This article is adapted from text provided by Kaustubh Wagh.







