New UMD Startup Builds the ‘Picks and Shovels’ for the RNA Revolution
Emergente’s patented RNAnneal platform predicts shapeshifting RNA structures, addressing a foundational bottleneck across the therapeutics, agriculture and biotechnology manufacturing industries.
A new University of Maryland startup takes aim at one of biotechnology’s most consequential infrastructure gaps: the inability to reliably design around RNA structure. RNA is rapidly emerging as a powerful means of intervention in biology—enabling therapies that silence, edit or activate genes beyond the reach of traditional protein-targeted drugs, including noncoding genomic regions implicated in disease.
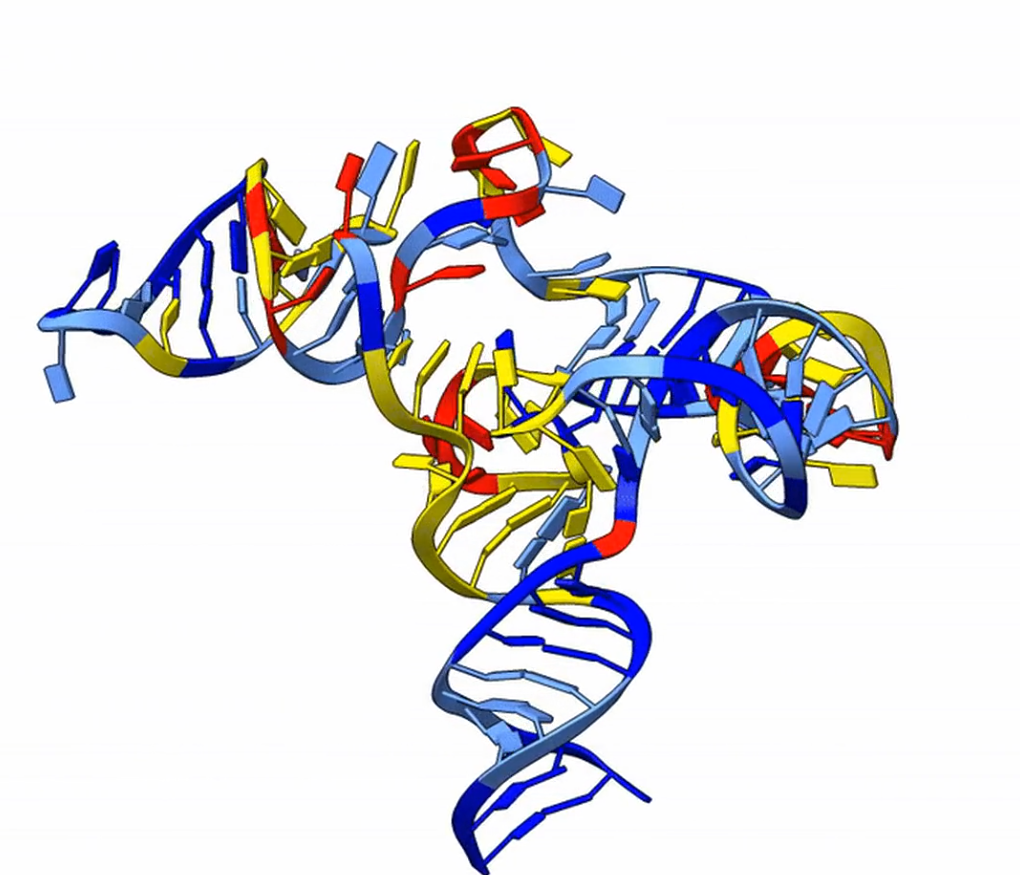
Designing RNAs and compounds that interact with them, however, is a difficult task. RNA molecules fold into diverse and dynamic three-dimensional shapes that are persistently difficult to predict and interpret. That structural uncertainty has become a major bottleneck for the field.
The College Park, Md.-based startup Emergente was formed to remove that bottleneck. Rather than building a single therapeutic asset, the company is developing foundational infrastructure that its co-founders call the “picks and shovels” powering the RNA economy. Its patented RNAnneal platform combines statistical physics, molecular simulation and artificial intelligence (AI) to generate thousands of structural ensembles from an RNA sequence and identify stable folds, ligand-binding pockets and functional dynamics. This approach shifts the paradigm from predicting a single static structure to mapping the full structural landscape governing biological behavior.
By directly addressing RNA’s structural variability, the platform has a wide range of therapeutic development applications, including oncology, neurology, genetic disease and infectious disease research. Its relevance also extends beyond human health to agriculture and animal biology, where RNA-targeting strategies are emerging as tools to address pathogen, pest and resilience challenges.
“The RNA bottleneck is not about discovery—millions of RNAs have been discovered—it is about design. Without structures, the RNA industry is operating in the dark, and the landscape of potential structures is a proverbial haystack. The tools we developed allow us to predict 3D structures, alternative folds, ligand-binding pockets and interactions for RNAs. They let us find the hidden needle,” said Emergente co-founder Pratyush Tiwary, a professor of chemistry and biochemistry at UMD, who also has appointments in the Institute for Physical Science and Technology at UMD, where he holds the Millard and Lee Alexander Professor in Chemical Physics; in the Department of Biochemistry and Molecular Biology at the University of Maryland, Baltimore; and at the University of Maryland Institute for Health Computing, where he leads the Center for Therapeutics Discovery.
Tiwary co-founded the company with UMD biophysics Ph.D. student Lukas Herron and Nadia Sarfraz, a Ph.D. RNA biochemist who recently graduated from Georgetown University. Following the development of the core technology in Tiwary’s lab, which received UMD’s 2025 Life Sciences Invention of the Year Award, the team licensed the patented intellectual property from the university and formed Emergente to commercialize RNAnneal. The technology also received competitive funding through a Maryland Innovation Initiative Technology Assessment Award.
Unlike many data-hungry modeling approaches that rely on vast structural training libraries, “RNAnneal leverages physics-grounded modeling, which makes it reliable even for previously uncharacterized RNA classes,” Herron said.
The platform uses principles of statistical physics, molecular simulations and AI to identify realistic structural configurations from the vast array of options. It also distinguishes between rigid and flexible parts of the molecule and does so very quickly: Users can upload a sequence to the platform hosted on the cloud and receive a full ensemble of structures back in less than 24 hours at a computational cost under $10. Comparable high-resolution cryogenic electron microscopy structure determination can cost upward of $100,000, while purely AI-based predictors such as AlphaFold3 do not reliably solve the analogous RNA structural-ensemble problem, according to Anne Simon, a professor of cell biology and molecular genetics at UMD and co-founder of Silvec Biologics.
Early benchmarking of RNAnneal on a class of RNAs called riboswitches demonstrated strong agreement with experimentally observed structures and competitive ranking performance against widely used modeling approaches.
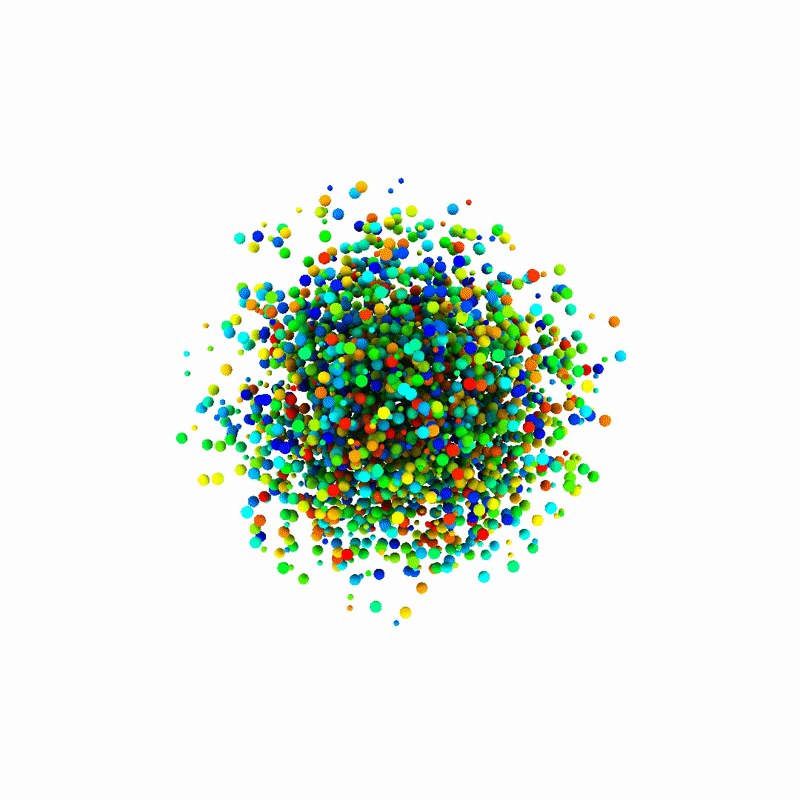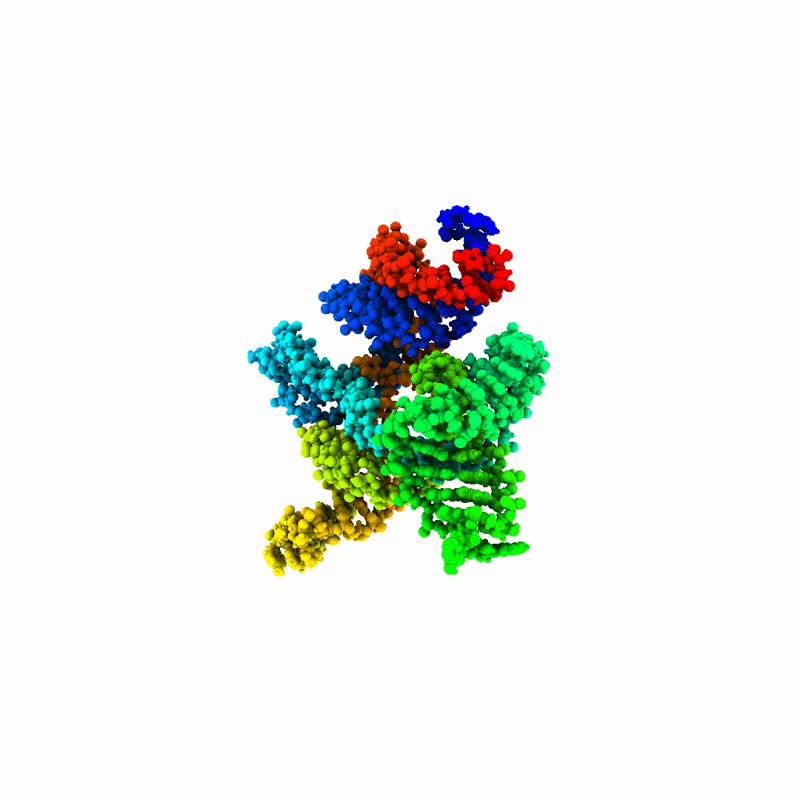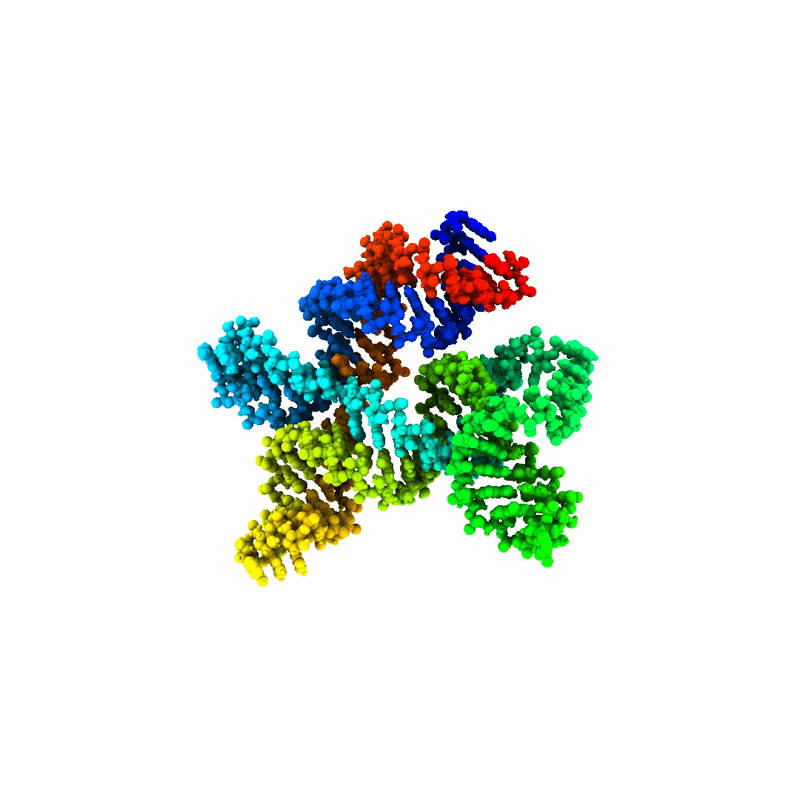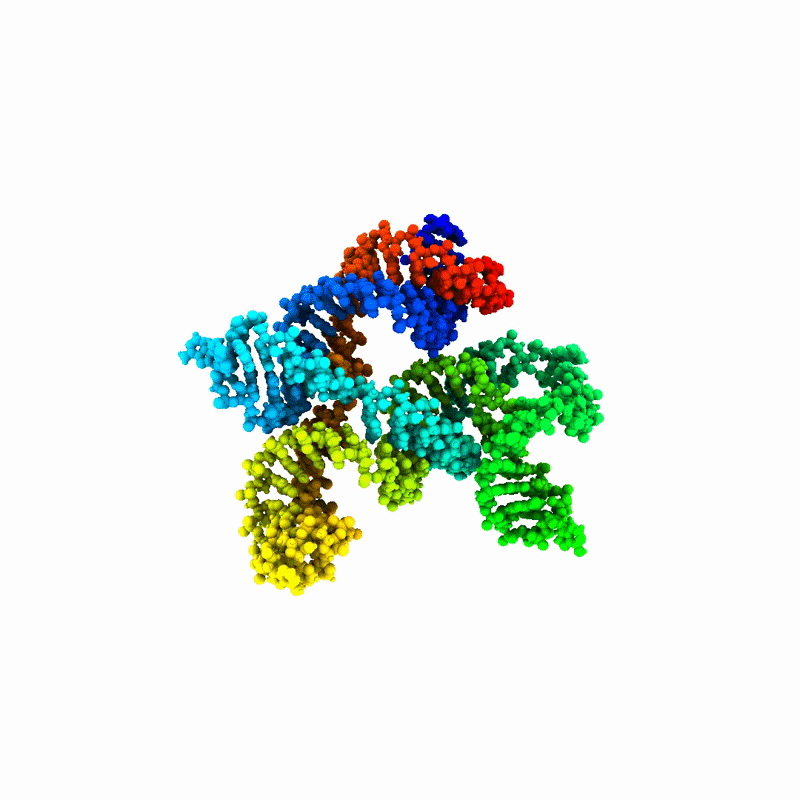
“RNA is the pasta of molecules. It can be spaghetti, penne or fusilli, and it’s constantly shape-shifting,” Tiwary explained. “That makes it very difficult to make predictions about it or build useful models.”
RNAnneal reflects a shift in focus, Tiwary said, from predicting a single structure to mapping an entire structural landscape. This focus highlights how RNA structure, regardless of which noodle it resembles, relates to function, disease and therapeutics.
“Biology is entering an era where success depends less on generating more data and more on extracting meaning from rare but valuable signals,” Tiwary said. “Our technology opens a scientific bottleneck, letting us into a previously inaccessible world that will lead to faster discoveries and better RNA-based technologies.”
The company's name is an ode to Nobel Prize-winning physicist Philip Anderson, who pioneered an emergent view of science: simple rules of interaction can lead to complex collective behavior. RNAs are built from just four letters—A, C, G and U—but from these few symbols and the simple rules of how they pair and stack emerges a vast landscape of shapes, dynamics and function.
###
This research has been funded by the National Institutes of Health’s National Institute of General Medical Sciences (Award No. R35GM142719), the U.S. National Science Foundation (Award No. CHE-2044165) and a Maryland Innovation Initiative Technology Assessment Award. This article does not necessarily reflect the views of these organizations.







