New Molecule Shows Promise in HIV Vaccine Design
Vaccine candidate spurs animals to produce antibodies against protective sugars of multiple HIV strains
Researchers at the University of Maryland and Duke University have designed a novel protein-sugar vaccine candidate that, in an animal model, stimulated an immune response against sugars that form a protective shield around HIV. The molecule could one day become part of a successful HIV vaccine.
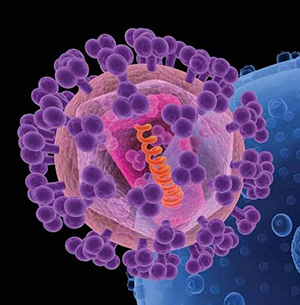 “An obstacle to creating an effective HIV vaccine is the difficulty of getting the immune system to generate antibodies against the sugar shield of multiple HIV strains,” said Lai-Xi Wang, a professor of chemistry and biochemistry at UMD. “Our method addresses this problem by designing a vaccine component that mimics a protein-sugar part of this shield.”
“An obstacle to creating an effective HIV vaccine is the difficulty of getting the immune system to generate antibodies against the sugar shield of multiple HIV strains,” said Lai-Xi Wang, a professor of chemistry and biochemistry at UMD. “Our method addresses this problem by designing a vaccine component that mimics a protein-sugar part of this shield.”
Wang and collaborators designed a vaccine candidate using an HIV protein fragment linked to a sugar group. When injected into rabbits, the vaccine candidate stimulated antibody responses against the sugar shield in four different HIV strains. The results were published in the journal Cell Chemical Biology on October 26, 2017.
The protein fragment of the vaccine candidate comes from gp120, a protein that covers HIV like a protective envelope. A sugar shield covers the gp120 envelope, bolstering HIV’s defenses. The rare HIV-infected individuals who can keep the virus at bay without medication typically have antibodies that attack gp120.
Researchers have tried to create an HIV vaccine targeting gp120, but had little success for two reasons. First, the sugar shield on HIV resembles sugars found in the human body and therefore does not stimulate a strong immune response. Second, more than 60 strains of HIV exist and the virus mutates frequently. As a result, antibodies against gp120 from one HIV strain will not protect against other strains or a mutant strain.
To overcome these challenges, Wang and his collaborators focused on a small fragment of gp120 protein that is common among HIV strains. The researchers used a synthetic chemistry method they previously developed to combine the gp120 fragment with a sugar molecule, also shared among HIV strains, to mimic the sugar shield on the HIV envelope.
Next, the researchers injected the protein-sugar vaccine candidate into rabbits and found that the rabbits’ immune systems produced antibodies that physically bound to gp120 found in four dominant strains of HIV in circulation today. Injecting rabbits with a vaccine candidate that contained the protein fragment without the sugar group resulted in antibodies that primarily bound to gp120 from only one HIV strain.
“This result was significant because producing antibodies that directly target the defensive sugar shield is an important step in developing immunity against the target and therefore the first step in developing a truly effective vaccine,” Wang said.
Although the rabbits’ antibodies bound to gp120, they did not prevent live HIV from infecting cells. This result did not surprise Wang, who noted that it usually takes humans up to two years to build immunity against HIV and the animal study only lasted two months.
“We have not hit a home run yet,” Wang noted. “But the ability of the vaccine candidate to raise substantial antibodies against the sugar shield in only two months is encouraging; other studies took up to four years to achieve similar results. This means that our molecule is a relatively strong inducer of the immune response.”
The researchers’ next steps will be to conduct longer-term studies in combination with other vaccine candidates, hone in on what areas of gp120 the antibodies are binding to and determine how they can increase the antibodies’ effectiveness at neutralizing HIV.
###
Other study co-authors affiliated with the Department of Chemistry and Biochemistry at the University of Maryland include former postdoctoral associate Hui Cai, faculty assistant John Giddens, former postdoctoral associate Jared Orwenyo, assistant research scientist Qiang Yang and graduate student Roushu Zhang.
This work was supported by the National Institutes of Health (Award No. R01AI113896). The content of this article does not necessarily reflect the views of the organization.
The research paper, “Synthetic Three-Component HIV-1 V3 Glycopeptide Immunogens Induce Glycan-Dependent Antibody Responses,” Hui Cai et al., was published in the journal Cell Chemical Biology on October 26, 2017.
Media Relations Contact: Irene Ying, 301-405-5204, zying@umd.edu
University of Maryland
College of Computer, Mathematical, and Natural Sciences
2300 Symons Hall
College Park, MD 20742
www.cmns.umd.edu
@UMDscience
About the College of Computer, Mathematical, and Natural Sciences
The College of Computer, Mathematical, and Natural Sciences at the University of Maryland educates more than 7,000 future scientific leaders in its undergraduate and graduate programs each year. The college’s 10 departments and more than a dozen interdisciplinary research centers foster scientific discovery with annual sponsored research funding exceeding $150 million.