Molecular Tug-of-war Gives Cells Their Shape
UMD researchers explain how cells form subcellular patterns
In a new study, University of Maryland researchers have demystified the process by which cells receive their shape—and it all starts with a protein called actin.
Actin is a key component of the cytoskeleton that provides structure to cells, much like how our skeletons support our bodies. However, unlike our skeleton, the actin cytoskeleton is a highly malleable structure that can rapidly assemble and disassemble in response to biochemical and biophysical cues.
It is well known that actin can form both 3D spherical shell-like structures that protect cells from external pressure and 2D rings that modify intracellular functions. But whenever researchers tried to recreate these structures outside the cell, they almost always ended up with clusters of actin. No one knew why—until now.
The researchers used computer simulations to show that actin and its partner protein, myosin, engage in a tug-of-war, with myosin trying to trap actin in local clusters and actin attempting to flee. If actin wins, actin filaments escape myosin’s pulling force and spontaneously form rings and spherical shells. If myosin wins, the actin network collapses and forms dense clusters.
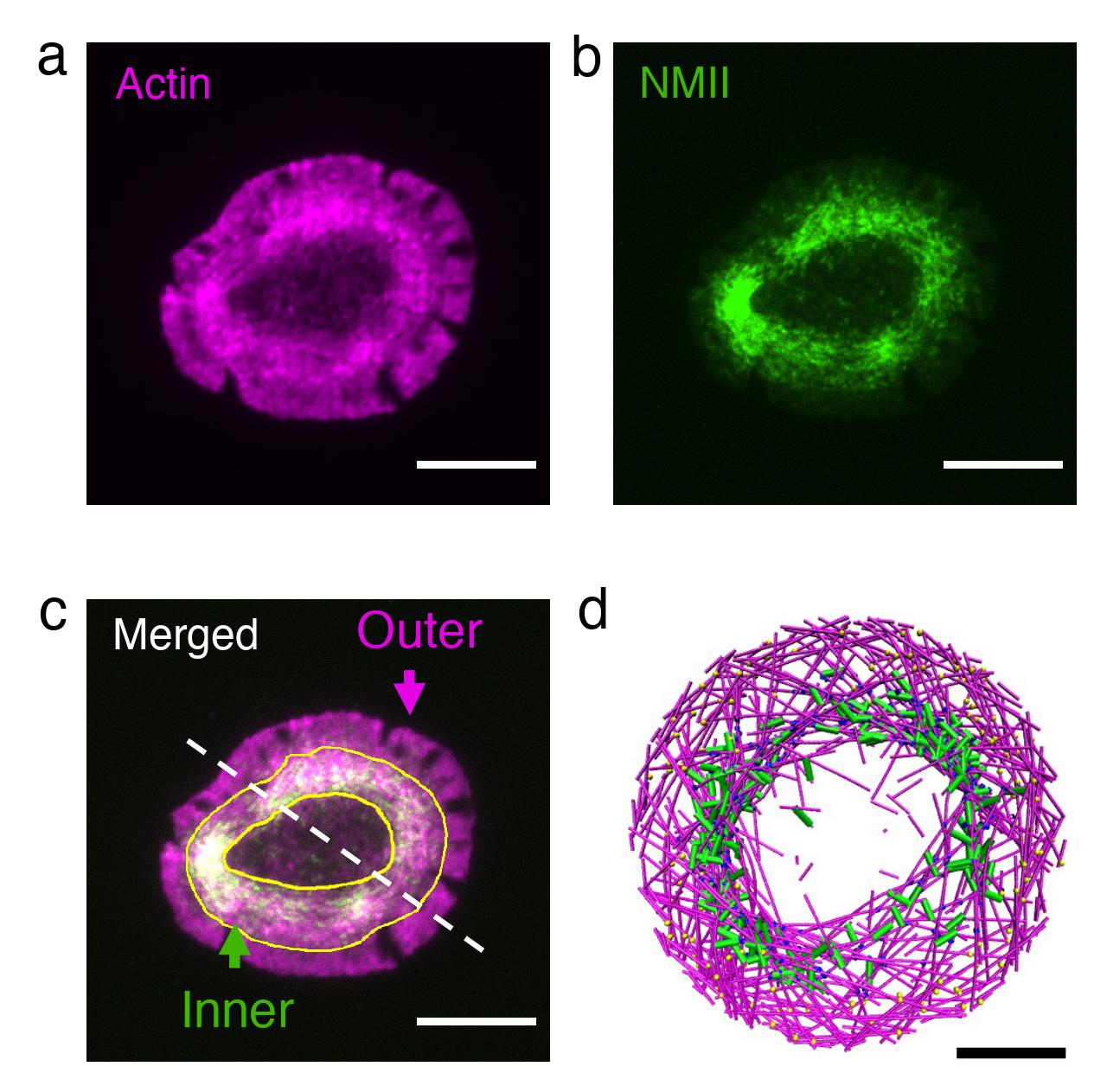
“Actin rings and spherical shells are ubiquitous in almost all cell types across species. We think that understanding the mechanism behind the formation of these structures unlocks the door to how cells sense and respond to their environment,” said Garegin Papoian, a co-author of the study and a UMD Monroe Martin Professor in the Department of Chemistry and Biochemistry and the Institute for Physical Science and Technology (IPST).
Their findings, published Oct. 21, 2022 in the journal eLife, could have important implications for human health. Because actin rings are central to our bodies’ ability to fight off foreign cells—with defects potentially resulting in impaired immunity or autoimmune disorders—the findings of this study could aid the development of future drugs.
Actin monomers can be thought of as railroad cars, which link up to form a train-like actin filament. These actin trains move through the cell because of a process called treadmilling. Also at play are the myosin motors, which pull oppositely oriented trains toward each other. Papoian, Qin Ni (Ph.D. ’21, chemical engineering) and biophysics Ph.D. student Haoran Ni believed that a competition between myosin’s pulling force and the rate of treadmilling was responsible for the formation of actin rings.
Fine-tuning these parameters in living cells is not possible, so the researchers turned to a simulation software called MEDYAN, developed by the Papoian Lab. MEDYAN uses physics and chemistry rules to simulate the dynamics of cytoskeletal proteins. They simulated an actin and myosin network (collectively referred to as actomyosin) in a thin disc and spherical shell.
They found that if the actin trains move slowly, the myosin pulling force causes traffic jams, which are the actomyosin clusters that have been observed in networks reconstituted outside cells. On the other hand, if the actin trains move fast, they can escape myosin’s pull. Once they reach the boundary of the disc, myosin’s pulling force makes the actin trains turn, preventing a head-on collision with the disc edge. Repeated occurrence of these events results in all the trains moving in a circle along the perimeter of the disc, which forms the actin ring.
Further analysis offers a thermodynamic theory to explain why cells form rings and shells. According to the laws of physics, systems favor the lowest energy configuration. Myosin proteins generate a lot of mechanical energy by bending actin filaments, which can only be released if actin can run away and relax. In living cells, actin’s ability to move fast enough to escape myosin and run to the edge allows for this built-up energy to be released, allowing for the formation of rings or shells, which, thermodynamically speaking, is the lowest energy configuration.
“The reason rings were not previously seen outside the cell is because actin just wasn’t moving fast enough,” Papoian said. “Myosin was winning 10 times out of 10.”
Together with Arpita Upadhyaya, a UMD physics professor with a joint appointment in IPST, and physics graduate student Kaustubh Wagh, biological sciences graduate student Aashli Pathni and biophysics graduate student Vishavdeep Vashisht, the team set out to test this model in living cells by turning their attention to T cells, where rings naturally form.
T cells are the cells in our body that hunt down foreign cells. When they recognize a cell as foreign and become activated, the T cell cytoskeleton rapidly reorganizes itself to form an actin ring at the cell-cell interface. Starting with cells that had formed rings, the researchers investigated the effect of perturbing actin and myosin using high-resolution live-cell imaging.
Reducing the actin train speed resulted in dissolution of the ring into small clusters, while increasing myosin’s pulling force led to rapid contraction of the ring, in remarkable agreement with associated simulations.
As a follow-up to this study, the team plans to add more complexity to the model and include other cytoskeletal components and organelles.
“We have been able to capture one fundamental aspect of cytoskeletal organization,” Papoian said. “Piece by piece, we plan to build a computational model of a complete cell using fundamental principles from physics and chemistry.”
###
This article is adapted from text provided by Qin Ni and Kaustubh Wagh.
The research paper, “A tug of war between filament treadmilling and myosin induced contractility generates actin rings,” was published in eLife on Oct. 21, 2022.
This work was supported by the National Science Foundation (Award Nos. CHE-1800418, PHY-1806903 and PHY-1607645) and the National Institutes of Health (Award No. R01 GM131054). This story does not necessarily reflect the views of these organizations.